sksfa.SFA¶
-
class
sksfa.SFA(n_components=None, *, batch_size=None, copy=True, svd_solver='full', tol=0.0, iterated_power='auto', random_state=None, robustness_cutoff=1e-15, fill_mode='noise')[source]¶ Slow Feature Analysis (SFA)
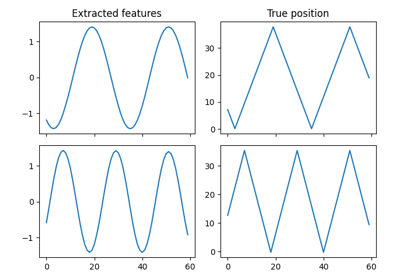
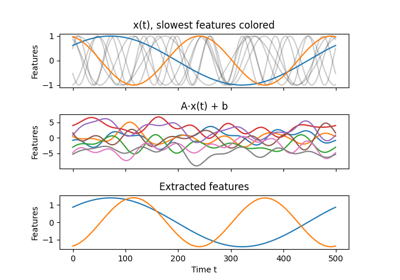
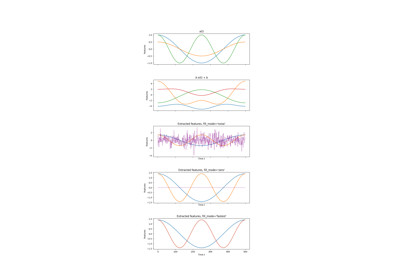
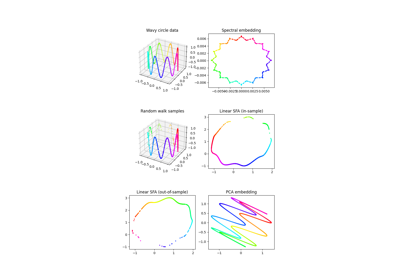
Linear dimensionality reduction and feature extraction method to be trained on time-series data. The data is decorrelated by whitening and linearly projected into the most slowly changing subspace. Slowness is measured by the average of squared one-step differences - thus, the most slowly changing subspacecorresponds to the directions with minimum variance for the dataset of one-step differences. It can be found using PCA.
After training, the reduction can be applied to non-timeseries data as well. Read more in the User Guide
- Parameters
- n_componentsint, float, None or str
Number of components to keep. If n_components is not set all components are kept.
- batch_sizeint or None
Batches of the provided sequence that should be considered individual time-series. If batch_size is not set, the whole sequence will be considered connected.
- copybool, default=True
If False, data passed to fit are overwritten and running fit(X).transform(X) will not yield the expected results, use fit_transform(X) instead.
- svd_solverstr {‘auto’, ‘full’, ‘arpack’, ‘randomized’}
The solver used by the internal PCA transformers. If auto :
The solver is selected by a default policy based on X.shape and n_components: if the input data is larger than 500x500 and the number of components to extract is lower than 80% of the smallest dimension of the data, then the more efficient ‘randomized’ method is enabled. Otherwise the exact full SVD is computed and optionally truncated afterwards.
- If full :
run exact full SVD calling the standard LAPACK solver via scipy.linalg.svd and select the components by postprocessing
- If arpack :
run SVD truncated to n_components calling ARPACK solver via scipy.sparse.linalg.svds. It requires strictly 0 < n_components < min(X.shape)
- If randomized :
run randomized SVD by the method of Halko et al.
- tolfloat >= 0, optional (default .0)
Tolerance for singular values computed by svd_solver == ‘arpack’ of the internal PCA transformers.
- iterated_powerint >= 0, or ‘auto’, (default ‘auto’)
Number of iterations for the power method computed by svd_solver == ‘randomized’.
- random_stateint, RandomState instance, default=None
Used when
svd_solver== ‘arpack’ or ‘randomized’. Pass an int for reproducible results across multiple function calls.- robustness_cutofffloat, default=1e-15
Applied after whitening. Features that have less explained variance in the input signal than robustness_cutoff will be considered trivial features and will be replaced in a way specified by fill_mode to ensure consistent output-dimensionality.
- fill_modestr {‘zero’, ‘fastest’, ‘noise’} or None (default ‘noise’)
The signal by which to replace output components that are artificial dimensions with almost no explained variance in input signal. If zero :
Output components will be replace with constant zero signals. Subsequent applications of SFA will pick this up as a trivial signal again.
- If fastest :
Output components will be replaced with copies of the fastest signals. Subsequent applications of SFA will pick this up as linearly dependent input dimension Since it is faster than any other signal, it would not be extracted in any case.
- If noise :
Output components will be replaced with independent streams of Gaussian noise. The streams are typically not heavily correlated, but are very fast and thus will not be extracted.
- If fill_mode is specifically set to None :
An exception will be thrown should trivial features are present in the input data.
Examples
>>> from sksfa import SFA >>> import numpy as np >>> >>> t = np.linspace(0, 8*np.pi, 1000).reshape(1000, 1) >>> t = t * np.arange(1, 6) >>> >>> ordered_cosines = np.cos(t) >>> mixed_cosines = np.dot(ordered_cosines, np.random.normal(0, 1, (5, 5))) >>> >>> sfa = SFA(n_components=2) >>> unmixed_cosines = sfa.fit_transform(mixed_cosines)
- Attributes
- input_dim_int
The number of input features.
- delta_values_array, shape (n_components,)
The estimated delta values (mean squared time-difference) of the different components.
- n_nontrivial_components_int
The number of components that did not fall under the threshold defined by robustness_cutoff. Read a: effective dimension of
input data.